Extract, transform & load: InfluxDB Cloud to Local PostgreSQL (Part 2)
Quick Recap
During part 1, I walked through the following actions in the effort to keep my cloud storage data for a longer duration:
- Burning a new OS image to a microSD
- Setting up a raspberry pi 3b+ to run headless
- Installing and configuring a postgresql server on the rpi
- Testing the postgresql instance by writing data to it remotely
This probably sounds backwards, if you did not read part 1, but to recap, I don’t pay for my InfluxDB cloud account and have a 30-day retention policy on my data storage. 30-days is fine for real-time monitoring but for longer term modeling, I have installed a postgreSQL database on an oldish raspberry pi 3b+ and aim to collect longintudinal health metrics. So, without further ado…
Extract, Transform and Load
First, I would like to offer an explanation as to why I have decided to not store the data on a local instance of InfluxDB and then I will give some insight into the data I have stored on the cloud, the structure of the data and the resolution. Finally, I will share the script I will be using to perform the ETL operation.
Why postgres rather than a local influxdb instance?
Using InfluxDB OSS, seemed like the most obvious choice. This was my initial intent. What stopped me short of doing this is the lack of an InfluxDB version >=2.0 available for my rpi3B+ architecture (armv7l). If I had a spare rpi4 lying around, this would have been my choice. I do have InfluxDB version 1.8.6-1 available in my repo:
influxdb/unknown,now 1.8.6-1 armhf
Distributed time-series database.
This version will allow me to query with both the legacy InfluxQL query language and Flux if I toggle that option within the configuration. But, I cannot write to the time-series database using Flux. I do not like the idea of being locked into the older version of InfluxDB and therefore, I chose postgres as my storage solution.
A glimpse of the raw data
The screenshot below was taken from InfluxData’s GUI Data Explorer. A few items to note. I did not aggregate the data. I take samples from my daughter’s pulse oximeter once a second. This is the same resolution I store in the cloud. The start time is the starting timeframe of the query (now() - 10s) in this case. The stop time is/was now(). The time column is the actual time of the sensor read. The value column is the sensor reading itself and is paired with the field column. The field in time-series lingo is an un-indexed column. The measurement column is analogous to the table name in a relational database which in this case is spo2 (not the best choice and I should have named it differently). Finally, UID is a tag or an indexed column. This is residual from me experimenting with multiple sensors and completely lacks any information or utility at this point-in-time.
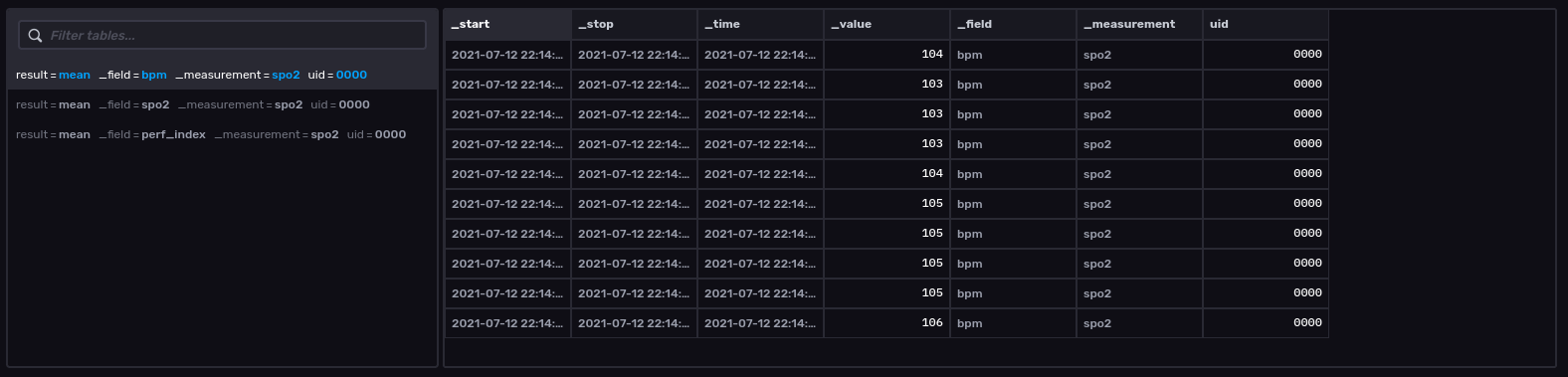
The sensor reads are all stored to InfluxDB in UTC time. I plan on doing the same for my postgresql instance. From my experience, timestamps can cause some real headaches and the clearest path is to store in UTC time and to handle any timezones on the application end while modeling.
ETL Script
So, a brief outline of what the script should do:
- Create a time range/constraint of one hour since I will be taking snapshots every hour of everyday
- Query InfluxDB using those created time constraints
- Handle as much of the transformation as I can during the query, e.g. Aggregate, Pivot and Drop columns
- Save query return to pandas dataframe
- Drop final un-wanted columns, convert timestamp into datetime64 and make dttm an index
- Push dataframe to postgresql instance, appending onto table if it exists
This script will only execute once, therefore, since I am taking one hour snapshots, I need to schedule the script to execute once per hour. More on that shortly.
# Filename: health_metrics_loader.py
# Author: Mike Hinkle
# Purpose: Extract data from InfluxDB cloud account, sanitize
# and push to local postgres instance
import os
import pandas as pd
from influxdb_client import InfluxDBClient
from sqlalchemy import create_engine
from dotenv import load_dotenv
from datetime import datetime, timedelta
load_dotenv()
#
# Load secrets from dotenv file
#
TOKEN = os.getenv("TOKEN")
ORG = os.getenv("ORG")
URL = os.getenv("CONNECTION")
PG_CONNECT = os.getenv("PG_CONNECT")
#
# Create time range to bound query
#
now = datetime.utcnow().strftime("%Y-%m-%dT%H:00:00Z")
last = datetime.utcnow() - timedelta(hours=1)
last_hour = last.strftime("%Y-%m-%dT%H:00:00Z")
#
# Instantiate connection object for TSDB
#
client = InfluxDBClient(url=URL, token=TOKEN, org=ORG)
query_api = client.query_api()
#
# Query InfluxDB cloud instance and return in df
#
df_current_sats = query_api.query_data_frame(
'from(bucket: "pulse_oximeter") '
"|> range(start: " + last_hour + ", stop: " + now + ") "
'|> filter(fn: (r) => r._measurement == "spo2") '
"|> aggregateWindow(every: 5s, fn: mean, createEmpty: false)"
'|> drop(columns: ["_start","_stop","_measurement","uid"])'
'|> pivot(rowKey:["_time"], columnKey: ["_field"], valueColumn: "_value")'
)
#
# Close connection object
#
client.__del__()
#
# Try and except to handle case when cloud provider is offline or
# rpi unplugged (away from wifi).
#
# Drop don't care columns, rename time header and set time as index
#
try:
df_current_sats.drop(labels=["table", "result"], axis=1, inplace=True)
df_current_sats.rename(columns={"_time": "time"}, inplace=True)
df_current_sats["time"] = pd.to_datetime(df_current_sats["time"])
df_current_sats["time"] = df_current_sats["time"].astype("datetime64[us]")
df_current_sats.set_index("time", inplace=True)
#
# Instantiate sqlalchemy connection object
#
engine = create_engine(PG_CONNECT)
#
# Use pandas to write dataframe to postgres instance
#
df_current_sats.to_sql("pox_five_second_mean", engine, if_exists="append")
#
# Close sqlalchemy connection object
#
engine.dispose()
except KeyError:
pass
Scheduling considerations
Now that the script is complete, and I know that it works, I need to ensure that it is run once per hour. The path of least resistence since I am running a single script on the same raspberry hosting the postgresql server, will be to run the script as a cronjob. I realize that there are more robust scheduling solutions such as Apache Airflow and I intend to do the same with a DAG (Directed Acyclic Graph) at a later date. For now, I want to get the data collected and I do not have airflow installed on the rpi3b+. So, forgive my rush and the simplicity of the solution.
After pushing the script to the raspberry pi 3b+, installing the dependencies (python-dotenv, psycopg2, sqlalchemy, influxdb-client and pandas) and creating my .env file containing my secrets, I can create the cronjob like so:
pi@influxdb-historic:~ $ crontab -e
0 * * * * /usr/bin/python3 /home/pi/pg_loader/health_metrics_loader.py >> ~/cron.log 2>&1
That should do the trick. I will check later tonight, plug my new postgres instance into my Grafana install running on my workstation and see if the data is collecting successfully.
8-hours later (Update)
It looks like the script and the cronjob were a success:
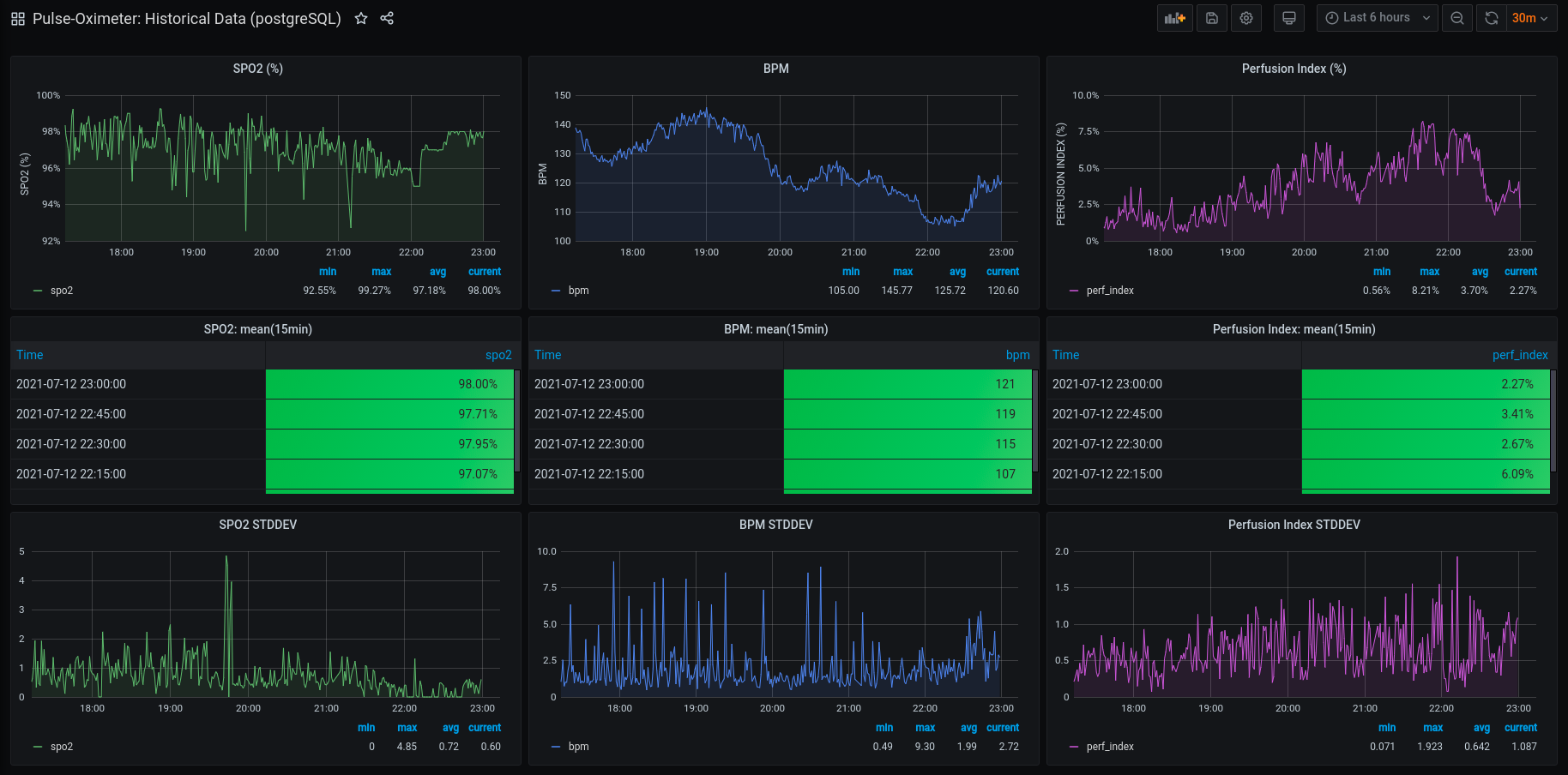
I will be revisiting this project to discuss various models when I have ample historical data. I might also redo the scheduling using Apache Airflow. I am hesitant since it seems like bird hunting with a scud missile and slightly overkill. If you see any opportunities for improvements or mistakes, please shoot me an email. Thank you for reading and for your time.
“Questions give us no rest.” ― Ayn Rand

Comments